Practical example: from raw gas concentration data to clean ecosystem gas fluxes.
fluxible_practical_example.RmdFor this example we will use the data that were recorded during the
Plant Functional Traits Course 6 (PFTC6) in Norway in 2022 at the site
called Liahovden. The
CO2 concentration data as well as air and soil temperature
and photosynthetically active radiations (PAR) were recorded in a
dataframe named co2_liahovden. The metadata for each
measurements are in a dataframe called record_liahovden.
This dataframe contains the starting time of each measurements, the type
of measurement and the unique ID for each turf. The type of measurement
describes if it was net ecosystem exchange (NEE), measured with a
transparent chamber, or ecosystem respiration (ER), measured with a dark
chamber.
We use the flux_match function to slice the data from
co2_liahovden into each measurement and discard what was
recorded in between.
library(fluxible)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(forcats)
str(record_liahovden)
#> tibble [138 × 3] (S3: tbl_df/tbl/data.frame)
#> $ turfID: chr [1:138] "4 AN1C 4" "4 AN1C 4" "27 AN3C 27" "27 AN3C 27" ...
#> $ type : chr [1:138] "NEE" "ER" "NEE" "ER" ...
#> $ start : POSIXct[1:138], format: "2022-07-27 05:37:30" "2022-07-27 05:42:00" ...
str(co2_liahovden)
#> tibble [89,692 × 5] (S3: tbl_df/tbl/data.frame)
#> $ datetime : POSIXct[1:89692], format: "2022-07-27 05:34:49" "2022-07-27 05:34:50" ...
#> $ temp_air : num [1:89692] 3 NA NA NA NA NA NA NA NA NA ...
#> $ temp_soil: num [1:89692] 2.96 NA NA NA NA NA NA NA NA NA ...
#> $ conc : num [1:89692] 468 469 468 468 468 ...
#> $ PAR : num [1:89692] 2.59 NA NA NA NA NA NA NA NA NA ...
conc_liahovden <- flux_match(
raw_conc = co2_liahovden,
field_record = record_liahovden,
startcrop = 0,
measurement_length = 220,
ratio_threshold = 0.5,
time_diff = 0,
datetime_col = "datetime",
conc_col = "conc",
start_col = "start"
)
# this exemple does not have a campaign column to pair the measurements
# we add pairID to pair NEE and ER for GEP calculation later
conc_liahovden <- conc_liahovden |>
mutate(
f_fluxID = as.double(f_fluxID),
pairID = case_when(
type == "NEE" ~ f_fluxID,
type == "ER" ~ f_fluxID - 1
),
f_fluxID = as_factor(f_fluxID),
pairID = as_factor(pairID)
)
str(conc_liahovden)
#> tibble [30,281 × 14] (S3: tbl_df/tbl/data.frame)
#> $ f_datetime : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:31" ...
#> $ temp_air : num [1:30281] NA NA NA NA NA NA NA NA NA 3 ...
#> $ temp_soil : num [1:30281] NA NA NA NA NA NA NA NA NA 6.83 ...
#> $ f_conc : num [1:30281] 468 467 467 467 467 ...
#> $ PAR : num [1:30281] NA NA NA NA NA ...
#> $ turfID : chr [1:30281] "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" ...
#> $ type : chr [1:30281] "NEE" "NEE" "NEE" "NEE" ...
#> $ f_start : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:30" ...
#> $ f_end : POSIXct[1:30281], format: "2022-07-27 05:41:10" "2022-07-27 05:41:10" ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_ratio : num [1:30281] 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_flag_match: chr [1:30281] NA NA NA NA ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 1 1 1 1 1 1 1 1 1 ...Before calculating fluxes we need to fit a model to each measurement
and estimate a slope of the concentration changing rate. We use the
flux_fitting function with the model provided by Zhao et al. (2018). The function
flux_fitting also provides a quadratic and a linear
fit.
slopes_exp_liahovden <- flux_fitting(
conc_df = conc_liahovden,
start_cut = 0,
end_cut = 0,
start_col = "f_start",
end_col = "f_end",
datetime_col = "f_datetime",
conc_col = "f_conc",
fluxid_col = "f_fluxID",
t_window = 20,
cz_window = 15,
b_window = 10,
a_window = 10,
roll_width = 15,
t_zero = 0,
fit_type = "exponential"
)
#> Cutting measurements...
#> Estimating starting parameters for optimization...
#> Optimizing fitting parameters...
#> Calculating fits and slopes...
#> Done.
#> Warning in flux_fitting_exp(conc_df, start_cut = ((start_cut)), end_cut = ((end_cut)), :
#> fluxID 77 : slope was estimated on 194 points out of 220 seconds
#> fluxID 81 : slope was estimated on 217 points out of 220 seconds
#> fluxID 83 : slope was estimated on 215 points out of 220 seconds
#> fluxID 85 : slope was estimated on 175 points out of 220 seconds
str(slopes_exp_liahovden)
#> tibble [30,281 × 31] (S3: tbl_df/tbl/data.frame)
#> $ f_datetime : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:31" ...
#> $ temp_air : num [1:30281] NA NA NA NA NA NA NA NA NA 3 ...
#> $ temp_soil : num [1:30281] NA NA NA NA NA NA NA NA NA 6.83 ...
#> $ f_conc : num [1:30281] 468 467 467 467 467 ...
#> $ PAR : num [1:30281] NA NA NA NA NA ...
#> $ turfID : chr [1:30281] "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" ...
#> $ type : chr [1:30281] "NEE" "NEE" "NEE" "NEE" ...
#> $ f_start : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:30" ...
#> $ f_end : POSIXct[1:30281], format: "2022-07-27 05:41:10" "2022-07-27 05:41:10" ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_ratio : num [1:30281] 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_flag_match: chr [1:30281] NA NA NA NA ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_time : num [1:30281] 0 1 2 3 4 5 6 7 8 9 ...
#> $ f_cut : Factor w/ 1 level "keep": 1 1 1 1 1 1 1 1 1 1 ...
#> $ n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ Cm_est : num [1:30281] 435 435 435 435 435 ...
#> $ a_est : num [1:30281] -0.249 -0.249 -0.249 -0.249 -0.249 ...
#> $ b_est : num [1:30281] -0.00288 -0.00288 -0.00288 -0.00288 -0.00288 ...
#> $ tz_est : num [1:30281] 14 14 14 14 14 14 14 14 14 14 ...
#> $ f_Cz : num [1:30281] 467 467 467 467 467 ...
#> $ time_diff : num [1:30281] 0 0 0 0 0 0 0 0 0 0 ...
#> $ f_Cm : num [1:30281] 372 372 372 372 372 ...
#> $ f_a : num [1:30281] -0.402 -0.402 -0.402 -0.402 -0.402 ...
#> $ f_b : num [1:30281] -0.00229 -0.00229 -0.00229 -0.00229 -0.00229 ...
#> $ f_tz : num [1:30281] 13.2 13.2 13.2 13.2 13.2 ...
#> $ f_slope : num [1:30281] -0.184 -0.184 -0.184 -0.184 -0.184 ...
#> $ f_fit : num [1:30281] 470 470 470 469 469 ...
#> $ f_fit_slope : num [1:30281] 470 470 470 469 469 ...
#> $ f_start_z : POSIXct[1:30281], format: "2022-07-27 05:37:43" "2022-07-27 05:37:43" ...
#> - attr(*, "fit_type")= chr "exponential"
slopes_qua_liahovden <- flux_fitting(
conc_df = conc_liahovden,
start_cut = 0,
end_cut = 0,
start_col = "f_start",
end_col = "f_end",
datetime_col = "f_datetime",
conc_col = "f_conc",
fluxid_col = "f_fluxID",
t_window = 20,
cz_window = 15,
b_window = 10,
a_window = 10,
roll_width = 15,
t_zero = 5,
fit_type = "quadratic"
)
#> Warning in flux_fitting_quadratic(conc_df, start_cut = ((start_cut)), end_cut = ((end_cut)), :
#> fluxID 77 : slope was estimated on 194 points out of 220 seconds because data are missing
#> fluxID 81 : slope was estimated on 217 points out of 220 seconds because data are missing
#> fluxID 83 : slope was estimated on 215 points out of 220 seconds because data are missing
#> fluxID 85 : slope was estimated on 175 points out of 220 seconds because data are missing
str(slopes_qua_liahovden)
#> tibble [30,281 × 26] (S3: tbl_df/tbl/data.frame)
#> $ f_datetime : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:31" ...
#> $ temp_air : num [1:30281] NA NA NA NA NA NA NA NA NA 3 ...
#> $ temp_soil : num [1:30281] NA NA NA NA NA NA NA NA NA 6.83 ...
#> $ f_conc : num [1:30281] 468 467 467 467 467 ...
#> $ PAR : num [1:30281] NA NA NA NA NA ...
#> $ turfID : chr [1:30281] "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" ...
#> $ type : chr [1:30281] "NEE" "NEE" "NEE" "NEE" ...
#> $ f_start : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:30" ...
#> $ f_end : POSIXct[1:30281], format: "2022-07-27 05:41:10" "2022-07-27 05:41:10" ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_ratio : num [1:30281] 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_flag_match : chr [1:30281] NA NA NA NA ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_time : num [1:30281] 0 1 2 3 4 5 6 7 8 9 ...
#> $ f_cut : Factor w/ 1 level "keep": 1 1 1 1 1 1 1 1 1 1 ...
#> $ n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_rsquared : num [1:30281] 0.967 0.967 0.967 0.967 0.967 ...
#> $ f_adj_rsquared: num [1:30281] 0.966 0.966 0.966 0.966 0.966 ...
#> $ f_pvalue : Named num [1:30281] 9.07e-161 9.07e-161 9.07e-161 9.07e-161 9.07e-161 ...
#> ..- attr(*, "names")= chr [1:30281] "value" "value" "value" "value" ...
#> $ f_intercept : num [1:30281] 465 465 465 465 465 ...
#> $ f_param1 : num [1:30281] -0.11 -0.11 -0.11 -0.11 -0.11 ...
#> $ f_param2 : num [1:30281] 3.9e-06 3.9e-06 3.9e-06 3.9e-06 3.9e-06 ...
#> $ f_slope : num [1:30281] -0.11 -0.11 -0.11 -0.11 -0.11 ...
#> $ f_fit : num [1:30281] 465 465 464 464 464 ...
#> $ f_fit_slope : num [1:30281] 465 465 464 464 464 ...
#> - attr(*, "fit_type")= chr "quadratic"
slopes_lin_liahovden <- flux_fitting(
conc_df = conc_liahovden,
start_cut = 0,
end_cut = 0,
start_col = "f_start",
end_col = "f_end",
datetime_col = "f_datetime",
conc_col = "f_conc",
fluxid_col = "f_fluxID",
t_window = 20,
cz_window = 15,
b_window = 10,
a_window = 10,
roll_width = 15,
t_zero = 5,
fit_type = "linear"
)
#> Warning in flux_fitting_lin(conc_df, start_cut = ((start_cut)), end_cut = ((end_cut)), :
#> fluxID 77 : slope was estimated on 194 points out of 220 seconds
#> fluxID 81 : slope was estimated on 217 points out of 220 seconds
#> fluxID 83 : slope was estimated on 215 points out of 220 seconds
#> fluxID 85 : slope was estimated on 175 points out of 220 seconds
str(slopes_lin_liahovden)
#> tibble [30,281 × 23] (S3: tbl_df/tbl/data.frame)
#> $ f_datetime : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:31" ...
#> $ temp_air : num [1:30281] NA NA NA NA NA NA NA NA NA 3 ...
#> $ temp_soil : num [1:30281] NA NA NA NA NA NA NA NA NA 6.83 ...
#> $ f_conc : num [1:30281] 468 467 467 467 467 ...
#> $ PAR : num [1:30281] NA NA NA NA NA ...
#> $ turfID : chr [1:30281] "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" ...
#> $ type : chr [1:30281] "NEE" "NEE" "NEE" "NEE" ...
#> $ f_start : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:30" ...
#> $ f_end : POSIXct[1:30281], format: "2022-07-27 05:41:10" "2022-07-27 05:41:10" ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_ratio : num [1:30281] 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_flag_match : chr [1:30281] NA NA NA NA ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_time : num [1:30281] 0 1 2 3 4 5 6 7 8 9 ...
#> $ f_cut : Factor w/ 1 level "keep": 1 1 1 1 1 1 1 1 1 1 ...
#> $ n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_rsquared : num [1:30281] 0.967 0.967 0.967 0.967 0.967 ...
#> $ f_adj_rsquared: num [1:30281] 0.966 0.966 0.966 0.966 0.966 ...
#> $ f_pvalue : Named num [1:30281] 9.23e-163 9.23e-163 9.23e-163 9.23e-163 9.23e-163 ...
#> ..- attr(*, "names")= chr [1:30281] "value" "value" "value" "value" ...
#> $ f_intercept : num [1:30281] 465 465 465 465 465 ...
#> $ f_slope : num [1:30281] -0.109 -0.109 -0.109 -0.109 -0.109 ...
#> $ f_fit : num [1:30281] 465 465 464 464 464 ...
#> - attr(*, "fit_type")= chr "linear"The function flux_quality is used to provide diagnostics
about the quality of the fit, eventually advising to discard some
measurements or replace them by zero.
slopes_exp_liahovden_flag <- flux_quality(
slopes_df = slopes_exp_liahovden,
# fit_type is automatically provided as an attribute because
# slopes_exp_liahovden was produced with flux_fitting
ambient_conc = 421,
error = 100,
fluxid_col = "f_fluxID",
slope_col = "f_slope",
force_discard = c(),
force_ok = c(),
ratio_threshold = 0,
conc_col = "f_conc",
b_col = "f_b",
time_col = "f_time",
fit_col = "f_fit",
cut_col = "f_cut",
rmse_threshold = 25,
cor_threshold = 0.5,
b_threshold = 1,
cut_arg = "cut"
)
#>
#> Total number of measurements: 138
#>
#> discard 4 3 %
#> ok 130 94 %
#> zero 4 3 %
#> weird_flux 0 0 %
#> start_error 0 0 %
#> no_data 0 0 %
#> force_ok 0 0 %
str(slopes_exp_liahovden_flag)
#> tibble [30,281 × 39] (S3: tbl_df/tbl/data.frame)
#> $ f_datetime : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:31" ...
#> $ temp_air : num [1:30281] NA NA NA NA NA NA NA NA NA 3 ...
#> $ temp_soil : num [1:30281] NA NA NA NA NA NA NA NA NA 6.83 ...
#> $ f_conc : num [1:30281] 468 467 467 467 467 ...
#> $ PAR : num [1:30281] NA NA NA NA NA ...
#> $ turfID : chr [1:30281] "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" ...
#> $ type : chr [1:30281] "NEE" "NEE" "NEE" "NEE" ...
#> $ f_start : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:30" ...
#> $ f_end : POSIXct[1:30281], format: "2022-07-27 05:41:10" "2022-07-27 05:41:10" ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_ratio : num [1:30281] 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_flag_match : chr [1:30281] NA NA NA NA ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_time : num [1:30281] 0 1 2 3 4 5 6 7 8 9 ...
#> $ f_cut : Factor w/ 1 level "keep": 1 1 1 1 1 1 1 1 1 1 ...
#> $ n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ Cm_est : num [1:30281] 435 435 435 435 435 ...
#> $ a_est : num [1:30281] -0.249 -0.249 -0.249 -0.249 -0.249 ...
#> $ b_est : num [1:30281] -0.00288 -0.00288 -0.00288 -0.00288 -0.00288 ...
#> $ tz_est : num [1:30281] 14 14 14 14 14 14 14 14 14 14 ...
#> $ f_Cz : num [1:30281] 467 467 467 467 467 ...
#> $ time_diff : num [1:30281] 0 0 0 0 0 0 0 0 0 0 ...
#> $ f_Cm : num [1:30281] 372 372 372 372 372 ...
#> $ f_a : num [1:30281] -0.402 -0.402 -0.402 -0.402 -0.402 ...
#> $ f_b : num [1:30281] -0.00229 -0.00229 -0.00229 -0.00229 -0.00229 ...
#> $ f_tz : num [1:30281] 13.2 13.2 13.2 13.2 13.2 ...
#> $ f_slope : num [1:30281] -0.184 -0.184 -0.184 -0.184 -0.184 ...
#> $ f_fit : num [1:30281] 470 470 470 469 469 ...
#> $ f_fit_slope : num [1:30281] 470 470 470 469 469 ...
#> $ f_start_z : POSIXct[1:30281], format: "2022-07-27 05:37:43" "2022-07-27 05:37:43" ...
#> $ f_flag_ratio : chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_start_error : chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_cor_coef : num [1:30281] -0.983 -0.983 -0.983 -0.983 -0.983 ...
#> $ f_RMSE : num [1:30281] 2.25 2.25 2.25 2.25 2.25 ...
#> $ f_fit_quality : chr [1:30281] NA NA NA NA ...
#> $ f_correlation : chr [1:30281] "yes" "yes" "yes" "yes" ...
#> $ f_quality_flag: chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_slope_corr : num [1:30281] -0.184 -0.184 -0.184 -0.184 -0.184 ...
#> - attr(*, "fit_type")= chr "exponential"
slopes_qua_liahovden_flag <- flux_quality(
slopes_df = slopes_qua_liahovden,
# fit_type is automatically provided as an attribute because
# slopes_exp_liahovden was produced with flux_fitting
ambient_conc = 421,
error = 100,
fluxid_col = "f_fluxID",
slope_col = "f_slope",
force_discard = c(),
force_ok = c(),
ratio_threshold = 0,
pvalue_col = "f_pvalue",
rsquared_col = "f_rsquared",
pvalue_threshold = 0.3,
rsquared_threshold = 0.7,
conc_col = "f_conc",
time_col = "f_time",
fit_col = "f_fit",
cut_col = "f_cut",
cut_arg = "cut"
)
#>
#> Total number of measurements: 138
#>
#> discard 1 1 %
#> ok 86 62 %
#> zero 51 37 %
#> weird_flux 0 0 %
#> start_error 0 0 %
#> no_data 0 0 %
#> force_ok 0 0 %
str(slopes_qua_liahovden_flag)
#> tibble [30,281 × 30] (S3: tbl_df/tbl/data.frame)
#> $ f_datetime : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:31" ...
#> $ temp_air : num [1:30281] NA NA NA NA NA NA NA NA NA 3 ...
#> $ temp_soil : num [1:30281] NA NA NA NA NA NA NA NA NA 6.83 ...
#> $ f_conc : num [1:30281] 468 467 467 467 467 ...
#> $ PAR : num [1:30281] NA NA NA NA NA ...
#> $ turfID : chr [1:30281] "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" ...
#> $ type : chr [1:30281] "NEE" "NEE" "NEE" "NEE" ...
#> $ f_start : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:30" ...
#> $ f_end : POSIXct[1:30281], format: "2022-07-27 05:41:10" "2022-07-27 05:41:10" ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_ratio : num [1:30281] 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_flag_match : chr [1:30281] NA NA NA NA ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_time : num [1:30281] 0 1 2 3 4 5 6 7 8 9 ...
#> $ f_cut : Factor w/ 1 level "keep": 1 1 1 1 1 1 1 1 1 1 ...
#> $ n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_rsquared : num [1:30281] 0.967 0.967 0.967 0.967 0.967 ...
#> $ f_adj_rsquared: num [1:30281] 0.966 0.966 0.966 0.966 0.966 ...
#> $ f_pvalue : Named num [1:30281] 9.07e-161 9.07e-161 9.07e-161 9.07e-161 9.07e-161 ...
#> ..- attr(*, "names")= chr [1:30281] "value" "value" "value" "value" ...
#> $ f_intercept : num [1:30281] 465 465 465 465 465 ...
#> $ f_param1 : num [1:30281] -0.11 -0.11 -0.11 -0.11 -0.11 ...
#> $ f_param2 : num [1:30281] 3.9e-06 3.9e-06 3.9e-06 3.9e-06 3.9e-06 ...
#> $ f_slope : num [1:30281] -0.11 -0.11 -0.11 -0.11 -0.11 ...
#> $ f_fit : num [1:30281] 465 465 464 464 464 ...
#> $ f_fit_slope : num [1:30281] 465 465 464 464 464 ...
#> $ f_flag_ratio : chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_start_error : chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_quality_flag: chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_slope_corr : num [1:30281] -0.11 -0.11 -0.11 -0.11 -0.11 ...
#> - attr(*, "fit_type")= chr "quadratic"
slopes_lin_liahovden_flag <- flux_quality(
slopes_df = slopes_lin_liahovden,
# fit_type is automatically provided as an attribute because
# slopes_exp_liahovden was produced with flux_fitting
ambient_conc = 421,
error = 100,
fluxid_col = "f_fluxID",
slope_col = "f_slope",
force_discard = c(),
force_ok = c(),
ratio_threshold = 0,
pvalue_col = "f_pvalue",
rsquared_col = "f_rsquared",
pvalue_threshold = 0.3,
rsquared_threshold = 0.7,
conc_col = "f_conc",
time_col = "f_time",
fit_col = "f_fit",
cut_col = "f_cut",
cut_arg = "cut"
)
#>
#> Total number of measurements: 138
#>
#> discard 6 4 %
#> ok 52 38 %
#> zero 80 58 %
#> weird_flux 0 0 %
#> start_error 0 0 %
#> no_data 0 0 %
#> force_ok 0 0 %
str(slopes_lin_liahovden_flag)
#> tibble [30,281 × 27] (S3: tbl_df/tbl/data.frame)
#> $ f_datetime : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:31" ...
#> $ temp_air : num [1:30281] NA NA NA NA NA NA NA NA NA 3 ...
#> $ temp_soil : num [1:30281] NA NA NA NA NA NA NA NA NA 6.83 ...
#> $ f_conc : num [1:30281] 468 467 467 467 467 ...
#> $ PAR : num [1:30281] NA NA NA NA NA ...
#> $ turfID : chr [1:30281] "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" "4 AN1C 4" ...
#> $ type : chr [1:30281] "NEE" "NEE" "NEE" "NEE" ...
#> $ f_start : POSIXct[1:30281], format: "2022-07-27 05:37:30" "2022-07-27 05:37:30" ...
#> $ f_end : POSIXct[1:30281], format: "2022-07-27 05:41:10" "2022-07-27 05:41:10" ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_ratio : num [1:30281] 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_flag_match : chr [1:30281] NA NA NA NA ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 1 1 1 1 1 1 1 1 1 ...
#> $ f_time : num [1:30281] 0 1 2 3 4 5 6 7 8 9 ...
#> $ f_cut : Factor w/ 1 level "keep": 1 1 1 1 1 1 1 1 1 1 ...
#> $ n_conc : int [1:30281] 220 220 220 220 220 220 220 220 220 220 ...
#> $ f_rsquared : num [1:30281] 0.967 0.967 0.967 0.967 0.967 ...
#> $ f_adj_rsquared: num [1:30281] 0.966 0.966 0.966 0.966 0.966 ...
#> $ f_pvalue : Named num [1:30281] 9.23e-163 9.23e-163 9.23e-163 9.23e-163 9.23e-163 ...
#> ..- attr(*, "names")= chr [1:30281] "value" "value" "value" "value" ...
#> $ f_intercept : num [1:30281] 465 465 465 465 465 ...
#> $ f_slope : num [1:30281] -0.109 -0.109 -0.109 -0.109 -0.109 ...
#> $ f_fit : num [1:30281] 465 465 464 464 464 ...
#> $ f_flag_ratio : chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_start_error : chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_quality_flag: chr [1:30281] "ok" "ok" "ok" "ok" ...
#> $ f_slope_corr : num [1:30281] -0.109 -0.109 -0.109 -0.109 -0.109 ...
#> - attr(*, "fit_type")= chr "linear"The function flux_plot provides plots for a visual
assessment of the measurements, explicitly displaying the quality flags
from flux_quality and the cuts from
flux_fitting.
slopes_exp_liahovden_flag |>
# we just show a sample of the plots to avoid slowing down the example
dplyr::filter(f_fluxID %in% c(54, 95, 100, 101)) |>
flux_plot(
color_discard = "#D55E00",
color_cut = "#D55E00",
color_ok = "#009E73",
color_zero = "#CC79A7",
f_date_breaks = "1 min",
f_minor_breaks = "10 sec",
f_date_labels = "%e/%m \n %H:%M",
f_ylim_upper = 600,
f_ylim_lower = 300,
f_plotname = "plot_quality",
facet_wrap_args = list(
ncol = 2,
nrow = 2,
scales = "free"
),
y_text_position = 400,
print_plot = "FALSE",
output = "print_only",
cut_arg = "cut",
no_data_flag = "no_data"
)
#> Part of the fit will not be displayed
#> because f_ylim_upper is too low.
#> Part of the fit will not be displayed
#> because f_ylim_lower is too high.
#> Plotting in progress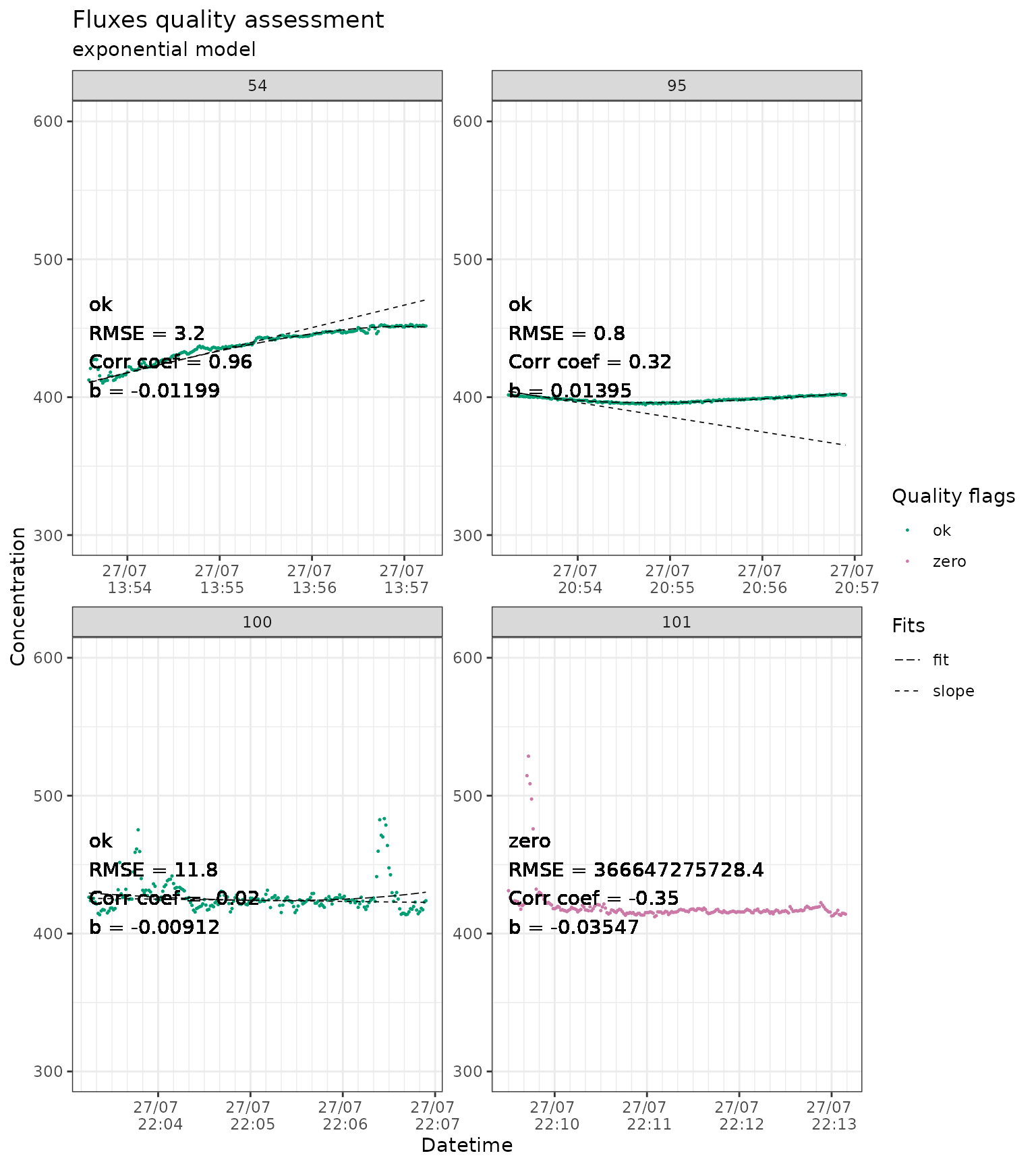
slopes_qua_liahovden_flag |>
# we just show a sample of the plots to avoid slowing down the example
dplyr::filter(f_fluxID %in% c(54, 95, 100, 101)) |>
flux_plot(
color_discard = "#D55E00",
color_cut = "#D55E00",
color_ok = "#009E73",
color_zero = "#CC79A7",
f_date_breaks = "1 min",
f_minor_breaks = "10 sec",
f_date_labels = "%e/%m \n %H:%M",
f_ylim_upper = 600,
f_ylim_lower = 300,
f_plotname = "plot_quality",
facet_wrap_args = list(
ncol = 2,
nrow = 2,
scales = "free"
),
y_text_position = 400,
print_plot = "FALSE",
output = "print_only",
cut_arg = "cut",
no_data_flag = "no_data"
)
#> Plotting in progress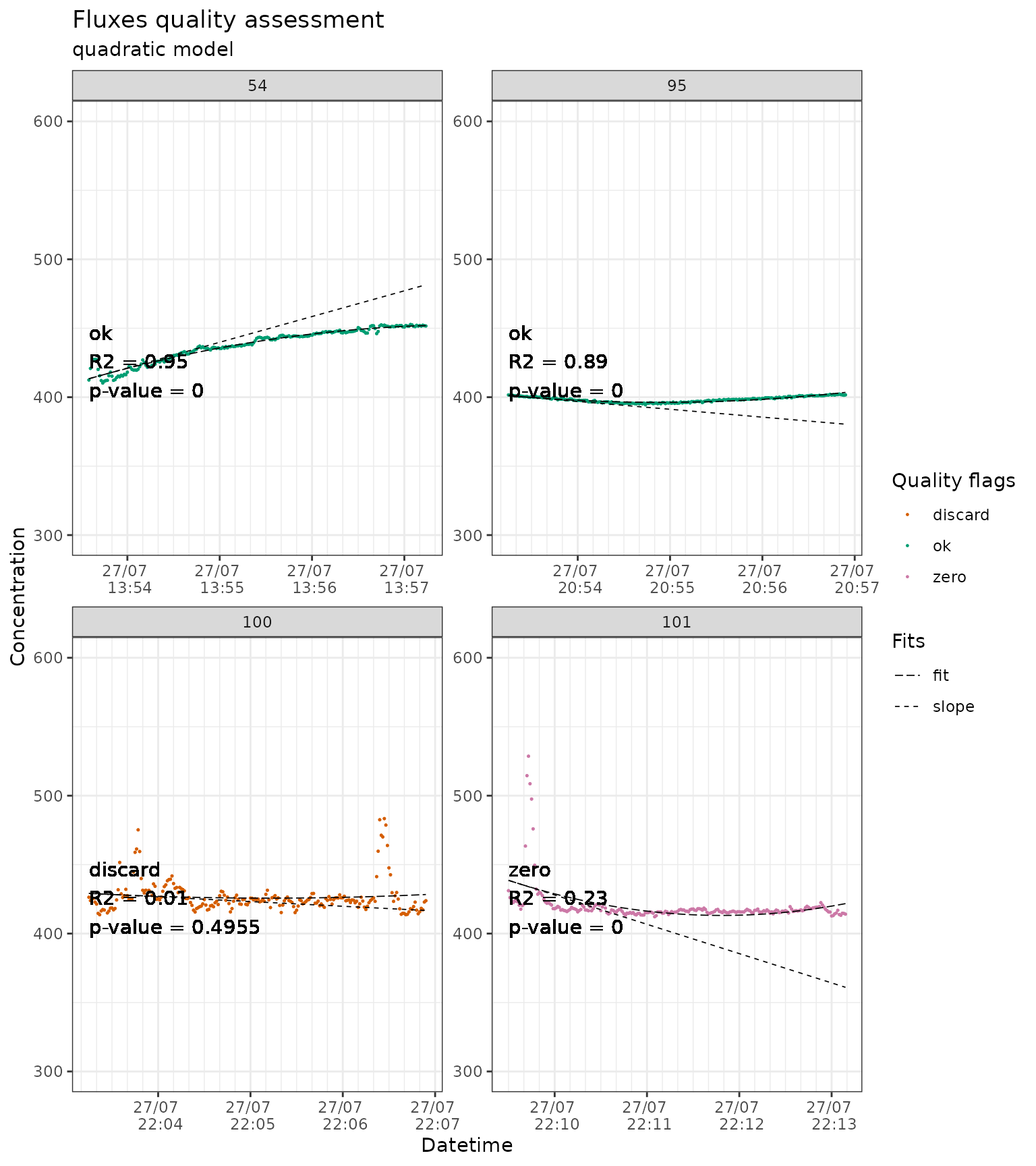
slopes_lin_liahovden_flag |>
# we just show a sample of the plots to avoid slowing down the example
dplyr::filter(f_fluxID %in% c(54, 95, 100, 101)) |>
flux_plot(
color_discard = "#D55E00",
color_cut = "#D55E00",
color_ok = "#009E73",
color_zero = "#CC79A7",
f_date_breaks = "1 min",
f_minor_breaks = "10 sec",
f_date_labels = "%e/%m \n %H:%M",
f_ylim_upper = 600,
f_ylim_lower = 300,
f_plotname = "plot_quality",
facet_wrap_args = list(
ncol = 2,
nrow = 2,
scales = "free"
),
y_text_position = 400,
print_plot = "FALSE",
output = "print_only",
cut_arg = "cut",
no_data_flag = "no_data"
)
#> Plotting in progress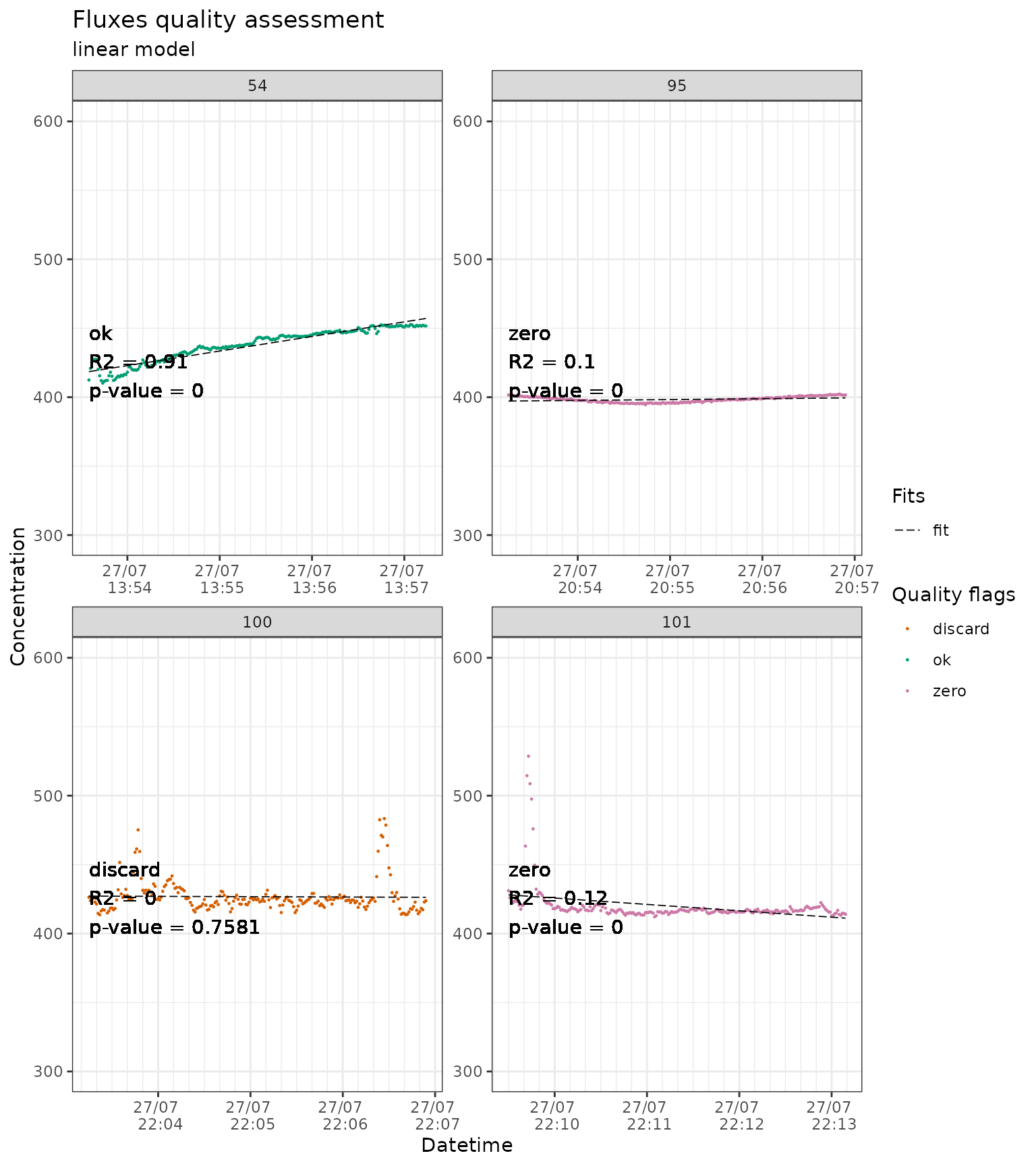
Based on the quality flags and the plots, the user can decide to run
flux_fitting again with different arguments. Here we will
do it while cutting the last 60 seconds of the fluxes (cutting the last
third). We also detected fluxes that do not look correct. Sometimes some
measurements will pass the automated quality control but are obviously
wrong for an experience user. That is what the
weird_fluxesID argument is for. For the sake of
reproducibility, this argument should be the last option and be
accompanied with a justification.
slopes_exp_liahovden_flag_60 <- conc_liahovden |>
flux_fitting(fit_type = "exp", end_cut = 60) |>
flux_quality(
slope_col = "f_slope",
force_discard = c(
51, # slope is much steeper than the flux because t zero was estimated
# at the very start of the measurement
101, # plot starts with a high peak: accumulation in the canopy?
106 # peak at the beginning of the flux that is messing up the fit
)
)
#> Cutting measurements...
#> Estimating starting parameters for optimization...
#> Optimizing fitting parameters...
#> Calculating fits and slopes...
#> Done.
#>
#> Total number of measurements: 135
#>
#> discard 1 1 %
#> ok 131 97 %
#> zero 3 2 %
#> weird_flux 0 0 %
#> start_error 0 0 %
#> no_data 0 0 %
#> force_ok 0 0 %
slopes_exp_liahovden_flag_60 |>
# we just show a sample of the plots to avoid slowing down the example
dplyr::filter(f_fluxID %in% c(54, 95, 100, 101)) |>
flux_plot(
f_ylim_lower = 300,
f_ylim_upper = 600,
facet_wrap_args = list(
ncol = 2,
nrow = 2,
scales = "free"
),
y_text_position = 400,
output = "print_only"
)
#> Part of the fit will not be displayed
#> because f_ylim_lower is too high.
#> Plotting in progress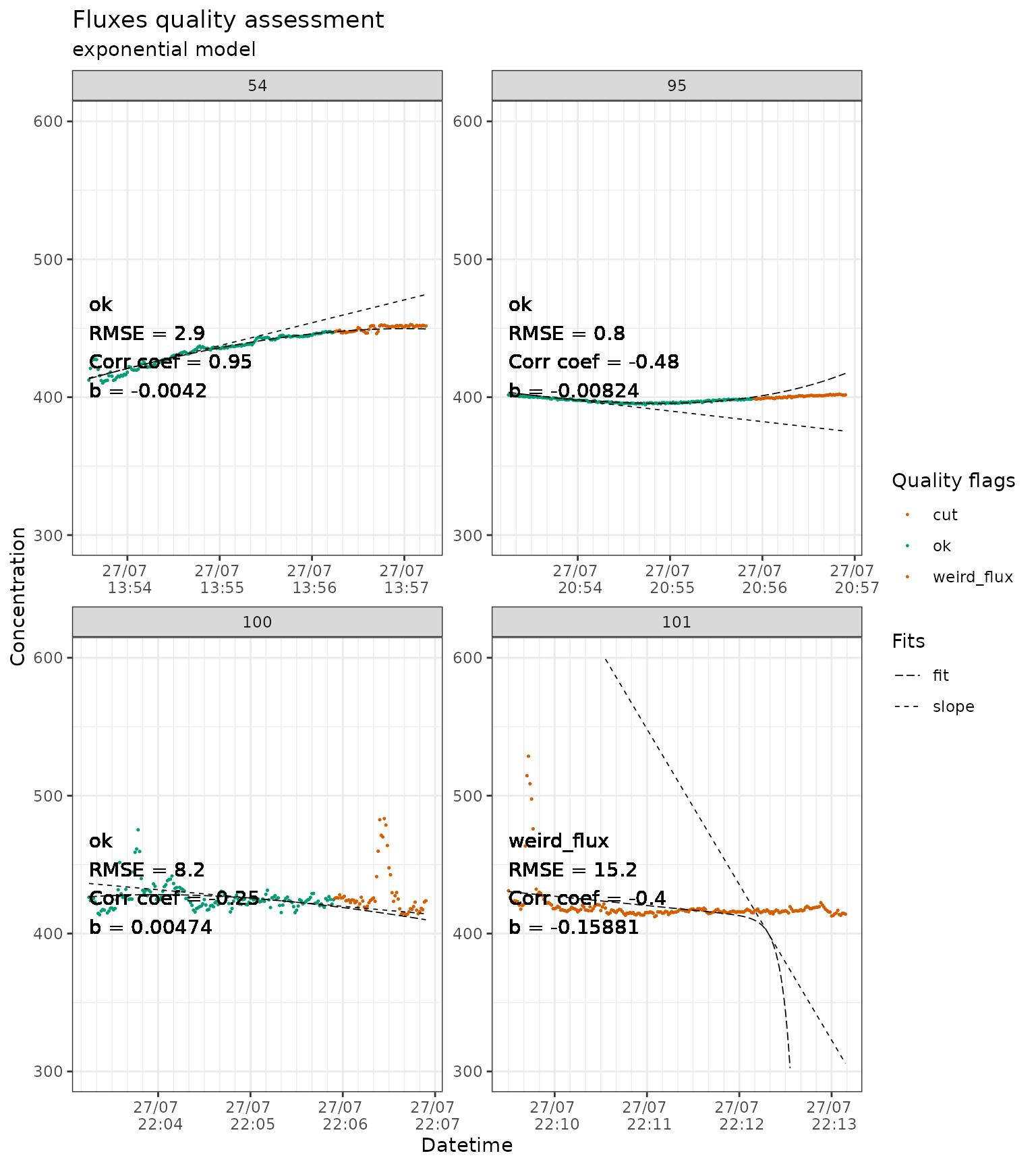
We also apply a cut on the dataset that was fitted with a quadratic model. At this point it is up to the user to decide which model works the best for the entire dataset. The function flux_quality provides a count of the quality flags that can help to take a decision.
slopes_qua_liahovden_flag_60 <- conc_liahovden |>
flux_fitting(fit_type = "qua", end_cut = 60, t_zero = 5) |>
flux_quality(
slope_col = "f_slope"
)
#>
#> Total number of measurements: 138
#>
#> ok 121 88 %
#> zero 17 12 %
#> discard 0 0 %
#> weird_flux 0 0 %
#> start_error 0 0 %
#> no_data 0 0 %
#> force_ok 0 0 %
slopes_qua_liahovden_flag_60 |>
# we just show a sample of the plots to avoid slowing down the example
dplyr::filter(f_fluxID %in% c(54, 95, 100, 101)) |>
flux_plot(
f_ylim_lower = 300,
f_ylim_upper = 600,
facet_wrap_args = list(
ncol = 2,
nrow = 2,
scales = "free"
),
y_text_position = 400,
output = "print_only"
)
#> Plotting in progress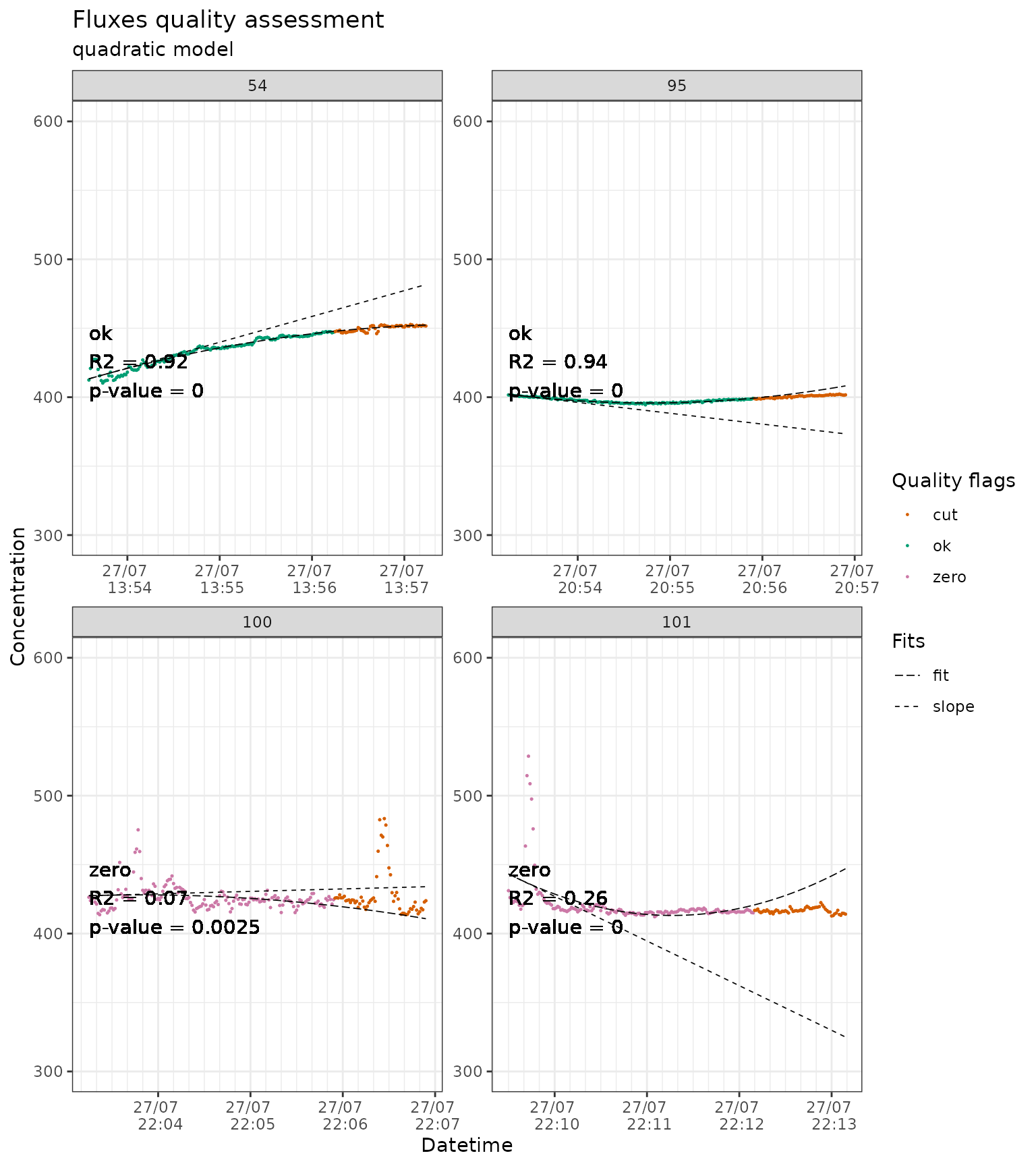
When using a linear fit it is common to take only a short section of the measurement close to the start. Here we will cut 120 seconds at the end, effectively keeping only the first 90 seconds.
slopes_lin_liahovden_flag_120 <- conc_liahovden |>
flux_fitting(fit_type = "lin", end_cut = 120, t_zero = 5) |>
flux_quality(
slope_col = "f_slope"
)
#>
#> Total number of measurements: 138
#>
#> discard 2 1 %
#> ok 109 79 %
#> zero 27 20 %
#> weird_flux 0 0 %
#> start_error 0 0 %
#> no_data 0 0 %
#> force_ok 0 0 %
slopes_lin_liahovden_flag_120 |>
# we just show a sample of the plots to avoid slowing down the example
dplyr::filter(f_fluxID %in% c(54, 95, 100, 101)) |>
flux_plot(
f_ylim_lower = 300,
f_ylim_upper = 600,
facet_wrap_args = list(
ncol = 2,
nrow = 2,
scales = "free"
),
y_text_position = 400,
output = "print_only"
)
#> Plotting in progress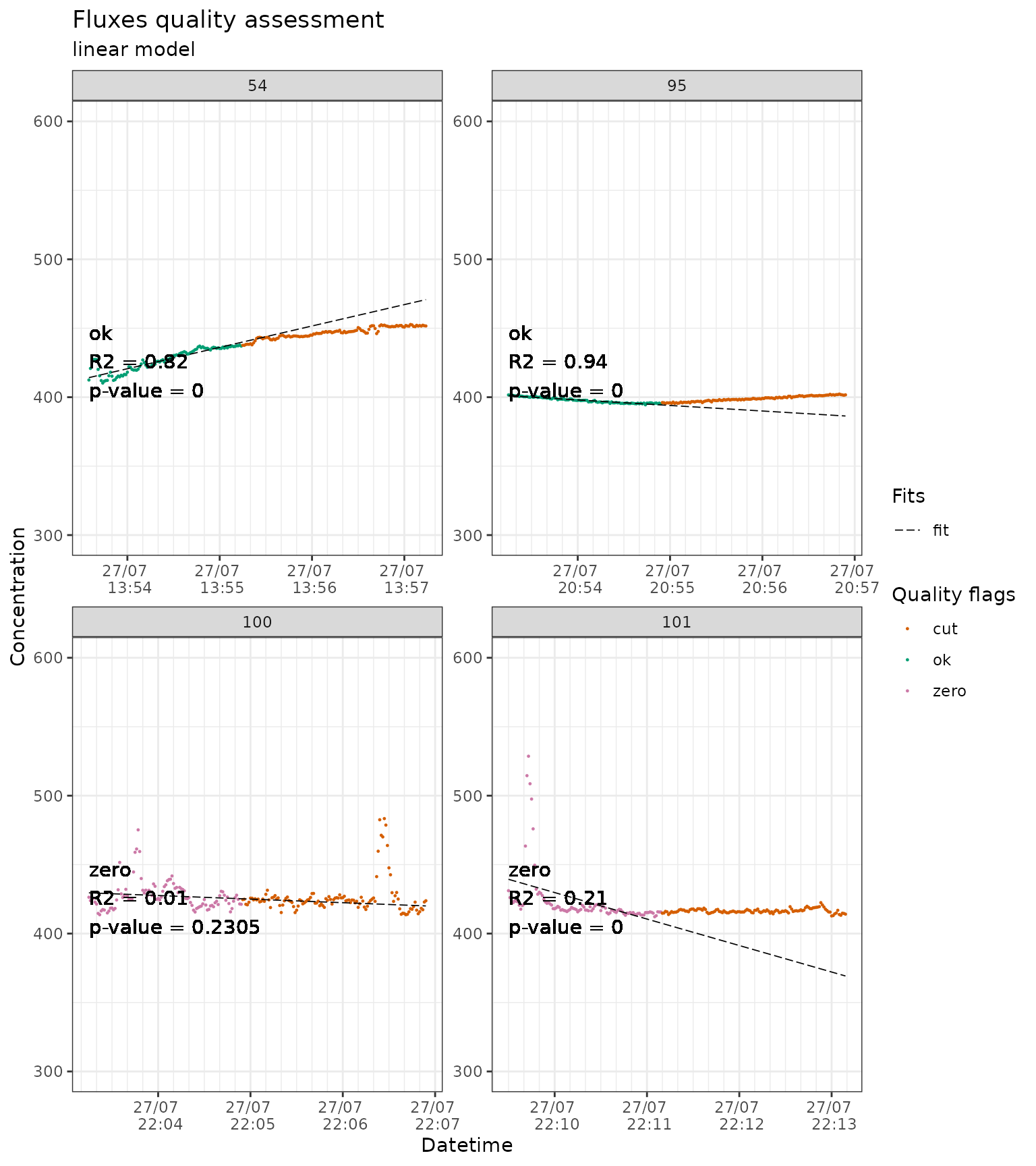
Once we are satisfied with the fit, we can calculate fluxes with
flux_calc. Here the volume is defined as a constant for all
the measurements but it is also possible to provide a specific volume
for each plot in case that is different.
fluxes_exp_liahovden_60 <- slopes_exp_liahovden_flag_60 |>
flux_calc(
slope_col = "f_slope_corr", # we use the slopes provided by flux_quality
datetime_col = "f_datetime",
conc_unit = "ppm",
flux_unit = "mmol",
cut_col = "f_cut",
keep_arg = "keep",
chamber_volume = 24.5,
tube_volume = 0.075,
atm_pressure = 1,
plot_area = 0.0625,
cols_keep = c("turfID", "type", "pairID"),
cols_ave = c("temp_soil", "PAR"),
fluxid_col = "f_fluxID",
temp_air_col = "temp_air",
temp_air_unit = "celsius"
)
#> Cutting data according to 'keep_arg'...
#> Averaging air temperature for each flux...
#> Creating a df with the columns from 'cols_keep' argument...
#> Creating a df with the columns from 'cols_ave' argument...
#> Calculating fluxes...
#> R constant set to 0.082057
#> Concentration was measured in ppm
#> Fluxes are in mmol/m2/h
fluxes_qua_liahovden_60 <- slopes_qua_liahovden_flag_60 |>
flux_calc(
slope_col = "f_slope_corr", # we use the slopes provided by flux_quality
datetime_col = "f_datetime",
conc_unit = "ppm",
flux_unit = "mmol",
cut_col = "f_cut",
keep_arg = "keep",
chamber_volume = 24.5,
tube_volume = 0.075,
atm_pressure = 1,
plot_area = 0.0625,
cols_keep = c("turfID", "type", "pairID"),
cols_ave = c("temp_soil", "PAR"),
fluxid_col = "f_fluxID",
temp_air_col = "temp_air",
temp_air_unit = "celsius"
)
#> Cutting data according to 'keep_arg'...
#> Averaging air temperature for each flux...
#> Creating a df with the columns from 'cols_keep' argument...
#> Creating a df with the columns from 'cols_ave' argument...
#> Calculating fluxes...
#> R constant set to 0.082057
#> Concentration was measured in ppm
#> Fluxes are in mmol/m2/h
fluxes_lin_liahovden_120 <- slopes_lin_liahovden_flag_120 |>
flux_calc(
slope_col = "f_slope_corr", # we use the slopes provided by flux_quality
datetime_col = "f_datetime",
conc_unit = "ppm",
flux_unit = "mmol",
cut_col = "f_cut",
keep_arg = "keep",
chamber_volume = 24.5,
tube_volume = 0.075,
atm_pressure = 1,
plot_area = 0.0625,
cols_keep = c("turfID", "type", "pairID"),
cols_ave = c("temp_soil", "PAR"),
fluxid_col = "f_fluxID",
temp_air_col = "temp_air",
temp_air_unit = "celsius"
)
#> Cutting data according to 'keep_arg'...
#> Averaging air temperature for each flux...
#> Creating a df with the columns from 'cols_keep' argument...
#> Creating a df with the columns from 'cols_ave' argument...
#> Calculating fluxes...
#> R constant set to 0.082057
#> Concentration was measured in ppm
#> Fluxes are in mmol/m2/hThe output is in mmol/m2/h and the calculation used is as follow:
where
flux: the flux of gas at the surface of the plot (mmol/m2/h)
slope: slope estimate (ppm*s-1)
P: pressure, assumed (atm)
V: volume of the chamber and tubing (L)
R: gas constant (0.082057 L*atm*K-1*mol-1)
T: chamber air temperature (K)
A: area of chamber frame base (m2)
The conversion from micromol/m2/s to mmol/m2/h is included in the function.
Fluxes were calculated in five steps from raw gas concentration data and the process is entirely reproducible.
Next we can calculated gross ecosystem production (GEP) and plot the results:
fluxes_exp_liahovden_60_gep <- fluxes_exp_liahovden_60 |>
flux_gep(
id_cols = "pairID",
flux_col = "flux",
type_col = "type",
datetime_col = "datetime",
par_col = "PAR",
cols_keep = c("temp_soil", "model", "turfID")
)
str(fluxes_exp_liahovden_60_gep)
#> tibble [207 × 16] (S3: tbl_df/tbl/data.frame)
#> $ datetime : POSIXct[1:207], format: "2022-07-27 05:37:30" "2022-07-27 05:46:55" ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 2 3 4 5 6 7 8 9 10 ...
#> $ PAR : num [1:207] 24.2 28.8 48.1 161.5 121.4 ...
#> $ type : chr [1:207] "GEP" "GEP" "GEP" "GEP" ...
#> $ flux : num [1:207] -18.57 -38.42 -20.18 -5.9 -6.99 ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: NA NA NA NA NA NA NA NA NA NA ...
#> $ temp_soil : num [1:207] 6.96 6.83 2.5 6.99 6.7 ...
#> $ turfID : chr [1:207] "4 AN1C 4" "27 AN3C 27" "77 AN2C 77" "4 AN1C 4" ...
#> $ f_slope_calc : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ chamber_volume: num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ tube_volume : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ atm_pressure : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ plot_area : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ temp_air_ave : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ volume_setup : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ model : chr [1:207] "exponential" "exponential" "exponential" "exponential" ...
fluxes_qua_liahovden_60_gep <- fluxes_qua_liahovden_60 |>
flux_gep(
id_cols = "pairID",
flux_col = "flux",
type_col = "type",
datetime_col = "datetime",
par_col = "PAR",
cols_keep = c("temp_soil", "model", "turfID")
)
str(fluxes_qua_liahovden_60_gep)
#> tibble [207 × 16] (S3: tbl_df/tbl/data.frame)
#> $ datetime : POSIXct[1:207], format: "2022-07-27 05:37:30" "2022-07-27 05:46:55" ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 2 3 4 5 6 7 8 9 10 ...
#> $ PAR : num [1:207] 24.2 28.8 48.1 161.5 121.4 ...
#> $ type : chr [1:207] "GEP" "GEP" "GEP" "GEP" ...
#> $ flux : num [1:207] -13.53 -32.5 -10.35 -6.55 -6.79 ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: NA NA NA NA NA NA NA NA NA NA ...
#> $ temp_soil : num [1:207] 6.96 6.83 2.5 6.99 6.7 ...
#> $ turfID : chr [1:207] "4 AN1C 4" "27 AN3C 27" "77 AN2C 77" "4 AN1C 4" ...
#> $ f_slope_calc : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ chamber_volume: num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ tube_volume : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ atm_pressure : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ plot_area : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ temp_air_ave : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ volume_setup : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ model : chr [1:207] "quadratic" "quadratic" "quadratic" "quadratic" ...
fluxes_lin_liahovden_120_gep <- fluxes_lin_liahovden_120 |>
flux_gep(
id_cols = "pairID",
flux_col = "flux",
type_col = "type",
datetime_col = "datetime",
par_col = "PAR",
cols_keep = c("temp_soil", "model", "turfID")
)
str(fluxes_lin_liahovden_120_gep)
#> tibble [207 × 16] (S3: tbl_df/tbl/data.frame)
#> $ datetime : POSIXct[1:207], format: "2022-07-27 05:37:30" "2022-07-27 05:46:55" ...
#> $ pairID : Factor w/ 69 levels "1","3","5","7",..: 1 2 3 4 5 6 7 8 9 10 ...
#> $ PAR : num [1:207] 23 28.2 46 164.4 120.5 ...
#> $ type : chr [1:207] "GEP" "GEP" "GEP" "GEP" ...
#> $ flux : num [1:207] -10.02 -20.55 0 -3.29 -6.24 ...
#> $ f_fluxID : Factor w/ 138 levels "1","2","3","4",..: NA NA NA NA NA NA NA NA NA NA ...
#> $ temp_soil : num [1:207] 6.93 6.82 2.55 7 6.68 ...
#> $ turfID : chr [1:207] "4 AN1C 4" "27 AN3C 27" "77 AN2C 77" "4 AN1C 4" ...
#> $ f_slope_calc : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ chamber_volume: num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ tube_volume : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ atm_pressure : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ plot_area : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ temp_air_ave : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ volume_setup : num [1:207] NA NA NA NA NA NA NA NA NA NA ...
#> $ model : chr [1:207] "linear" "linear" "linear" "linear" ...
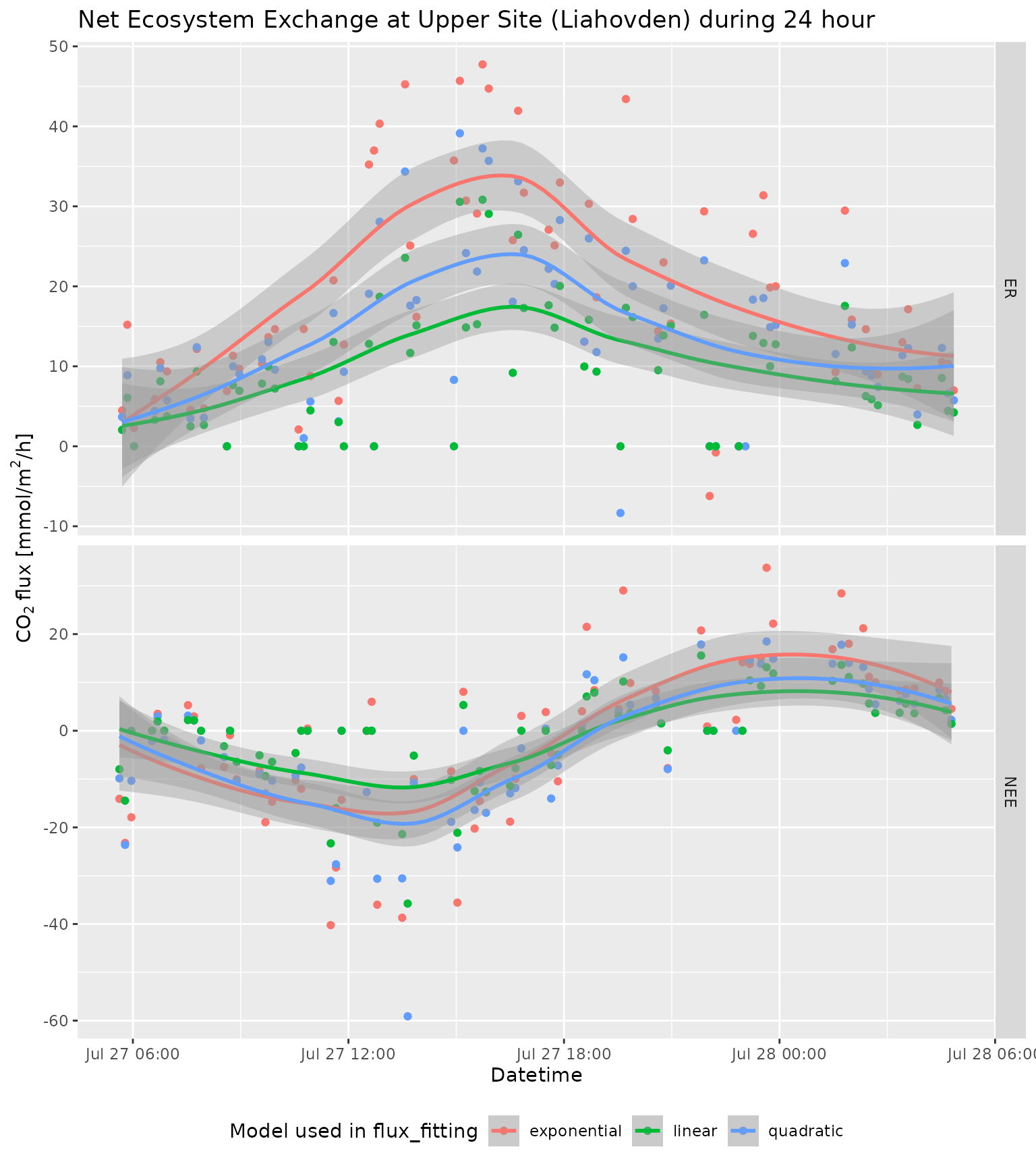
library(ggplot2)
bind_rows(
fluxes_exp_liahovden_60_gep,
fluxes_qua_liahovden_60_gep,
fluxes_lin_liahovden_120_gep
) |>
ggplot(aes(x = datetime, y = flux, color = model)) +
geom_point() +
geom_smooth() +
labs(
title = "Net Ecosystem Exchange at Upper Site (Liahovden) during 24 hour",
x = "Datetime",
y = bquote(~ CO[2] ~ "flux [mmol/" * m^2 * "/h]"),
color = "Model used in flux_fitting"
) +
theme(legend.position = "bottom") +
facet_grid(type ~ ., scales = "free")